Bernese TB Outbreak
Propagation of the Bernese Mycobacterium tuberculosis outbreak strain from 1991 over 20 years and its genomic microevolution, Switzerland (KEK 051/12)
One of the first molecular epidemiological TB studies in Switzerland identified a TB outbreak of 22 cases in the Canton of Berne 1991-1992 (Genewein et al., Lancet 1993). In 2012, we studied the molecular epidemiology of tuberculosis in Switzerland (see main project). We genotyped 520 M. tuberculosis isolates from patients diagnosed with tuberculosis between 2000 and 2008 (12.3% of all culture-confirmed tuberculosis cases in Switzerland during the study period). Among 68 isolates from the Canton of Bern, two were identified as belonging to the same outbreak described in 1993, indicating that this particular strain was still circulating in the region. The aim of the present study is to revisit this historical outbreak in Berne by whole genome sequences, and to study the propagation, genomic microevolution and epidemiological context until today. This will provide novel insights into the transmission dynamics of a TB outbreak over 20 years, provide a genomic transmission framework and assess the usefulness of whole genome sequencing compared to traditional techniques. The project was approved by the Cantonal Ethics Committee in Bern, and by the "Federal Expert Commission on Confidentiality in Medical Research" (Federal Office of Public Health) which allows the use of patient data under specified circumstances (decision letter).
This is a collaborative project between the Institute of Social and Preventive Medicine (University of Bern), the "Kantonsarzamt" Bern, the Bernese Lung Association, the Institute for Infectious Diseases (IFIK, University of Bern), and the Swiss Tropical and Public Health Institute (Basel). The project is funded by the Bernese Lung Association.
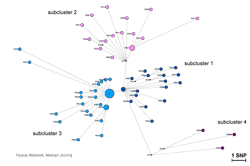
Strain-specific SNP-genotyping allowed rapid and inexpensive identification of M. tuberculosis outbreak isolates in a population-based strain collection. Subsequent targeted WGS provided detailed insights into transmission dynamics (Figure). This combined approach could be applied to track bacterial pathogens in real-time and at high resolution.
Publication: